CSIR - Centre for Cellular & Molecular Biology
Council of Scientific and Industrial Research
Ministry of Science & Technology, Govt. of India
Instructions for sample collection centres
Protocol to be followed at testing centres
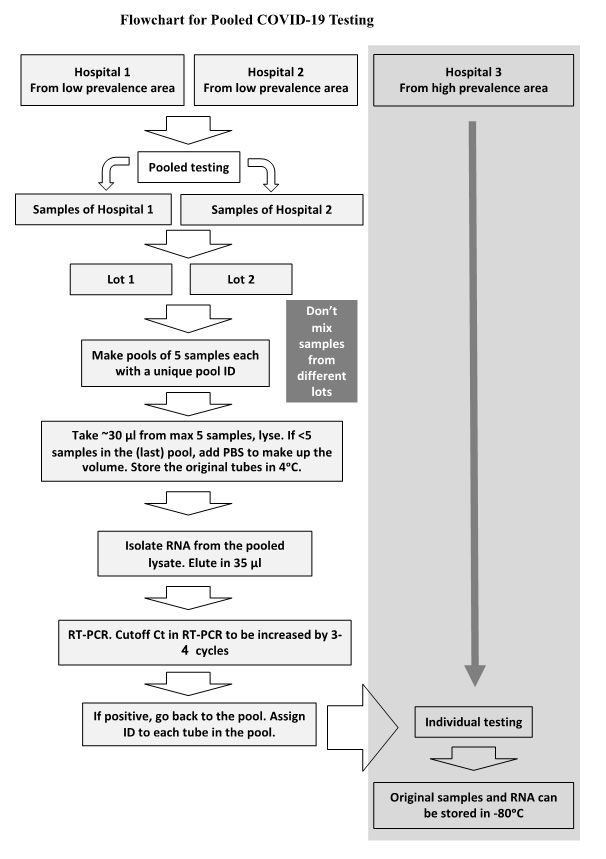
Samples will be received by the designated staff and parked in a designated place till processing for pooling starts.
The following description is for samples that are decided to be processed by pooling.
All individual samples that belonged to the negative pools will be reported as negative for SARS-CoV-2. The respective pool data (Ct value, etc.) will be associated with the five negative samples. Every single sample from the positive "pools" after re-testing individually will be reported as positive/negative, accordingly, with associated data (Ct value, etc.)