Guidance for converting academic research laboratory to
COVID-19 testing laboratory (operations, personnel, space organization and workflow)
Based on CCMB’s experience
Given its established research activities in viral pathogens and the availability of a BSL3 and BSL2+ facility, CCMB was the first academic laboratory to be designated as a testing centre for COVID-19, as well as the only non-ICMR lab to be designated as a validation centre for SARS-CoV-2 diagnostics. Following a thorough review of our readiness for performing these tasks safely and effectively, CCMB initiated testing on March 31, 2020. These SOPs incorporate WHO and ICMR best practices as outlined in the Handbook for COVID-19 testing issued by the PSA, Govt of India, and also provides additional details that might be of use to individual institutes. To date, CCMB has tested over 1000 samples.
Overall Operations: Coordinated by Director with a team of senior scientists and a larger advisory team
Personnel: Each step in the testing workflow has a designated coordinator. In addition to a designated team taking care of procurement, interactions with Govt hospitals, State Govt Health Department and CSIR HQ, there is a large group (30+) student volunteers who were trained in BSL3/2 operations.
Space organization: BSL3 and BSL2 facilities in Main Building for receiving samples, virus handling and sample aliquoting for RNA prep, additional BSL2 laboratory in iHUB where semiautomated RNA isolation and RT-PCR analysis are handled.
Workflow: is an optimized format developed for safety and convenience in our laboratory space.
CCMB scientists will be available to answer questions on any aspect of the operations and workflow for those laboratories that are planning these activities in the current pandemic.
Standard Operating Procedure for COVID-19 testing centre
April 15, 2020
These SOPs are governed by appropriate biosafety protocols and Personal Protection Equipment (PPE) at each stage.
- The State Government informs a designated person in the testing centre the number of samples they are sending.
- Samples arrive at CCMB in thermocol boxes brought by hospital ambulances.
Sample boxes are not to be removed from the ambulance at security office.
- The designated person at security confirms with the designated person who has been in touch with the State Government,if the samples are to be accepted or not.
- The designated person at the security checks the shipment papers, records (in a specific register for COVID-19 samples) the details of the samples (hospital that is sending the samples, when (date and time) it reaches the testing centre, and their number (number of boxes per case sheet, and number of case sheets). The sample boxes are not taken out of the ambulance.
- Based on the availability of volunteers in BSL facility the samples are to be taken directly to theBSL facility for testing or stored ina cold room at 4-6ᵒC. The designated security person accompanies the hospital personnel from security office to the appropriate location.
- The hospital personnel place the samples in the cold room or hand them to the volunteers at BSL facility along with their respective case sheets.
- If there is more than one box per shipment from a particular hospital, the security officer accompanying the hospital personnel ensures the boxes are numbered with marker so that the case sheets can be linked to all boxes from that hospital.
- Thestored samples are matched with the intimation received from the State Government by the designated person at the testing centre.
- The samples are then taken for documentation and testing in BSL facility.
- The test samples are taken into the BSL-3 lab inside the BSL facility, and the case sheets go to the COVID19 Documentation Room in the BSL facility.
- Every case sheet is allotted a unique code by the testing centre. This information is printed out on label stickers, and passed onto the volunteers in the BSL-3 lab who catalogue the samples received. They also receive a printout for records in a register in the BSL-3 lab.
- The name and age written on the sample tubes is visible to volunteers but not the area of its origin.
- The sample code number with its case sheet are cross-checked, and entered in a register in the BSL-3 lab.
- The sample packaging integrity (for tube leakage, labelling, cold chain) is checked, the sample tube surfaceis cleaned with alcohol (70% isopropanol), and are stored in -80ᵒC, in numbered racks until they need to be processed for testing.The rack number is also noted in the register along with other sample details.
- Upon their turn for testing, the samples are thawed and aliquoted. One of the aliquots is lysed, and set aside for viral RNA analysis. The code number of the sample aliquot is recorded in the register in the BSL-3 lab. The other aliquots of the same sample are stored in their designated locations in -80ᵒCfor future usage.
- The aliquots of each sample designated for testing, along with associated case sheets are taken to BSL-2 lab for testing.
- The sample lysates are received, verified and registered by the team in the COVID-19 BSL-2 testing facility.
- RNA is isolated from the samples in the RNA preparation room.
- The RNA is taken to the RT-PCR room for analysis.
- The RT-PCR pre-mix is prepared and distributed into 96-well plates in Hood #1. The plate is then shifted to Hood #2 where RNA template is added. The plate containing the complete reaction mix is then loaded into the RT-PCR machine. Readings are digitally recorded in realtime by the software in the associated computer. The file for each is named with catalogue number and date. These files are stored on the computer in datewise folders for collection by documentation team.
- After the reaction is complete the plate is discarded in a biohazard bag.
- Biohazard bags are autoclaved and disposed every day. The designated lab personnel for handling the biohazard bagswear the entire PPE kit, and move the biohazard bags into the Autoclave Room. After autoclaving at 121ᵒC for 20 min at a pressure of 15psa, these bags are sealed in cardboard boxes, and stored for being picked up by an external agency.
- Results from the computer attached to RT-PCR machine are downloaded in the pdf format onto pen drives, and pendrives carried to the documentation room by RT-PCR volunteers. The files are labelled with the date, batch and sample numbers, and arranged in a folder, date wise.
- The Data Analysis Team analyse the results. A sample is considered positive if the Ct value for a gene is determined to be<36. If a given sample tests positive for 2 out of 3 genes (genes E/N, ORF1b, RdRp), the case is considered to be COVID-19 positiveas per ICMRguidelines.
- The Analysis Team also decode the samples -thesample code allottedby the testing centre is matched with the case sheet of the patient received from the hospital.
- Entire patient details are filled in the testing centre’s portal.
- The test results are uploaded to the State Government portal, and emailed to pertinent State Government Officials, and also the respective hospitals where the samples originated from.
- The reports are also uploaded on the ICMR portal(https://cvstatus.icmr.gov.in).
- TheState Government is updated by the designated person at the testing centretwice a day by WhatsApp, at 7 am and 5 pm.
The case sheets are stored securely in the documentation room in the testing centre. All data is backed up on an external storage device. The data can only be accessed by authorized personnel.
- Biosafety protocols governing each location in sample receiving and handling workflow are as per guidelines.
- All personnel at each location have been trained to handle the samples according to the safety level required.
- Tables and floor of all labs are cleaned with 1% sodium hypochlorite.
- The sample boxes from the hospitals are cleaned with 1% sodium hypochlorite, followed by water and 70% isopropanol before opening them in BSL-3.
- Laminar hood areas are disinfected with UV exposure of 15 min daily, and cleaned with 70% ethanol/isopropanol.
- Testing and cleaning personnel clean their hands with Sterilium (45% isopropanol and 30% propanol).
- Entire PPE kits are worn during sample lysis, RNA isolation and RT-PCR in BSL-3 and BSL-2 labs.
- Below are the main roles which should have a primary person as well as a back up person.
| No |
Role |
|---|
| 1. | Connecting with state government |
|---|
| 2. | Pre-test documentation |
|---|
| 3. | BSL facility – oversee testing |
|---|
| 4. | Reagent and machine management |
|---|
| 5. | Data Analysis Team |
|---|
| 5. | Report generation |
|---|
- Roster for testingvolunteers
| No. |
Sample entry |
Batch 1 |
Batch 2 |
Frequency of turn of each batch |
| A |
Documentation |
A1, A2 |
A3, A4 |
Every day for half a day |
| B |
Sample storage and lysis |
B1, B2, B3 |
B4, B5, B6 |
Every day for half a day |
| C |
RNA isolation |
C1, C2 |
C5, C6 |
Every alternate day for half a day |
| C3, C4 |
C7, C8 |
| D |
RT-PCR |
D1, D2 |
D7, D8 |
Every alternate day for one third of a day |
| D3, D4 |
D9, D10 |
| D5, D6 |
D11, D12 |
| E |
Report generation |
E1, E2, E3 |
E4, E5, E6 |
Every day for half a day |
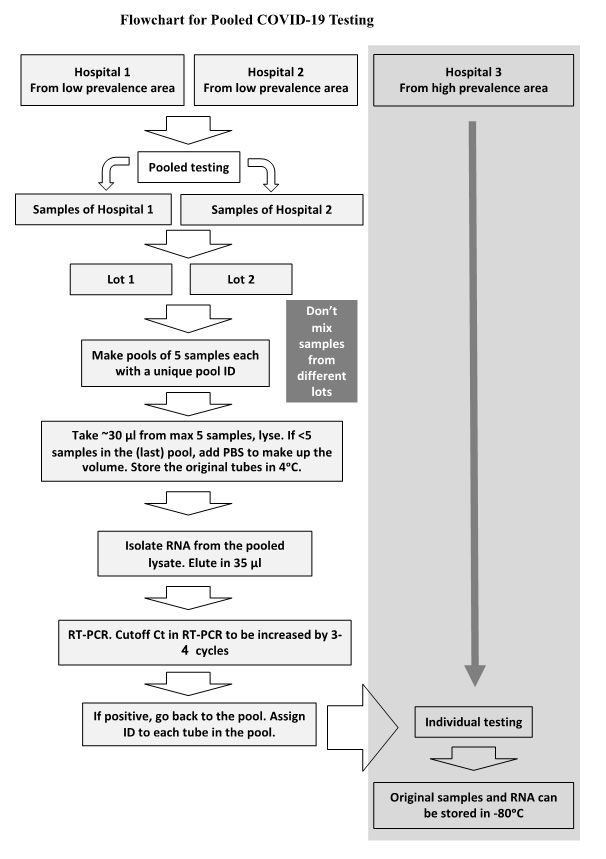
SOP for pooling of samples for COVID-19 testing by RT-PCR
Instructions for sample collection centres
- The packing of sample collection tubes should be as per ICMR-NIV guidelines. Parafilm over the lid + 1 absorbent layer + 2 plastic layers are sufficient, the plastic layers should be zip-lock bags. Please DO NOT over-pack or under-pack. No rubber band or cello tape.
- Kindly write the patient's name, age, gender (and other info, if required) legibly on the sample collection tube in such a way that the writing does not get erased during handling, use paper/cloth tape as label if needed and put cello tape over the labels.
- All samples should reach test centres within a fixed time window. It is important, as pooling starts, larger number of samples are required to start as a batch. Samples reaching after that time window will be delayed by 1 day.
- Pooling is useful when expected +ve is less than 4-5%. Therefore, an estimate/sample collection rationale will help pooling decision. Collection centers should be in contact with government health authorities to designate samples for pooling route of testing.
Protocol to be followed at testing centres
Samples will be received by the designated staff and parked in a designated place till processing for pooling starts.
The following description is for samples that are decided to be processed by pooling.
- Use the information in the case sheets to prepare an Excel spreadsheet that would contain the information for each sample provided in the case sheets. This is your "Sample Sheet" for a given Lot.
- Group these samples into separate hospitals/districts for every day separately, with the date and time of sample reception. Please use separate registers/notebooks for each hospital/district if you are not using computerized system. Label such groups as "Lot-1", "Lot-2", and so on. Within each "Lot", create bunch of five vials (picked randomly from the Lot) and label them as "Pool-1", "Pool-2", and so on.
- Pipette 30 µl of each sample (from a pool of 5) into a common 1.5 ml microcentrifuge tube to make a total of 150 µl. This tube has same label as that of respective pool. If the sample numbers are not in multiples of 5, combine 30 µl from each of the last 1/2/3/4 samples in the last pool and adjust the volume to 150 µl using 1x PBS.
- Immediately after completing one pool, secure the 5 tubes with a rubber band or put in a plastic beaker and label the pool. Ensure each pool stays as a discrete entity. Place multiple such pools in cardboard boxes and store them at 4°C fridge.
- Add lysis buffer to 150 µl of pooled samples.
- RNA isolation, reverse transcription, and real-time PCR: These steps proceed as usual, without any major changes because of the pooling. Elute in lesser volume (35 µl) to concentrate RNA.The Ct value cut-offs to classify a pool as negative or positive will need to be increased by 3-4 cycles due to dilution during pooling to ensure that sensitivity is not compromised.
- Once all qRT-PCRs are done, the pools that tested positive will be re-analysed by preparing RNA from individual samples of only those pools.
- Make a list of the unique identifiers of the samples in the pools that tested positive.
- Carry printouts of "Sample Sheet", the alphabetically-sorted patient details, of all the separate "Lots" into BSL-II/III lab.
- Take out only the pools that tested +ve from the fridge and now assign individual unique identifiers to each of those tubes, by marking on the printed list.
- Label the tubes and proceed with aliquoting and lysis as usual.
- Proceed with RNA isolation, reverse transcription and real-time PCR as usual.
- Store all the negative pools (with the existing rubber bands and labels) in a biohazard bag and keep inside BSL II/III lab until disinfected/autoclaved and discarded, as per the guidelines provided by the competent authority.
All individual samples that belonged to the negative pools will be reported as negative for SARS-CoV-2. The respective pool data (Ct value, etc.) will be associated with the five negative samples. Every single sample from the positive "pools" after re-testing individually will be reported as positive/negative, accordingly, with associated data (Ct value, etc.)